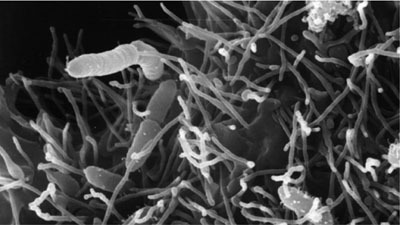
| Species: | Aeromonas veronii |
|---|---|
| Genus: | Aeromonas |
| Family: | Aeromonadaceae |
| Order: | Aeromonadales |
| Class: | Gammaproteobacteria |
| Phylum: | Proteobacteria |
| Shape | Rod-shaped |
|---|---|
| Gram staining | Gram- |
| Motility | Motile |
| Oxygen Requirement | Aerobe |
| Sporulation | Nonsporulating |
| Ecosystem | Annelida; Aquatic; Human; Fish |
| Ecosystem Type | Aquaculture; Circulatory system; Unclassified |
In the linked pathways:
red=enriched, blue=depleted
ko00010 - Glycolysis / Gluconeogenesis
ko00020 - Citrate cycle (TCA cycle)
ko00030 - Pentose phosphate pathway
ko00061 - Fatty acid biosynthesis
ko00071 - Fatty acid degradation
ko00072 - Synthesis and degradation of ketone bodies
ko00130 - Ubiquinone and other terpenoid-quinone biosynthesis
ko00230 - Purine metabolism
ko00240 - Pyrimidine metabolism
ko00250 - Alanine, aspartate and glutamate metabolism
ko00260 - Glycine, serine and threonine metabolism
ko00270 - Cysteine and methionine metabolism
ko00280 - Valine, leucine and isoleucine degradation
ko00281 - Geraniol degradation
ko00290 - Valine, leucine and isoleucine biosynthesis
ko00300 - Lysine biosynthesis
ko00330 - Arginine and proline metabolism
ko00340 - Histidine metabolism
ko00400 - Phenylalanine, tyrosine and tryptophan biosynthesis
ko00410 - beta-Alanine metabolism
ko00430 - Taurine and hypotaurine metabolism
ko00450 - Selenocompound metabolism
ko00471 - D-Glutamine and D-glutamate metabolism
ko00473 - D-Alanine metabolism
ko00480 - Glutathione metabolism
ko00500 - Starch and sucrose metabolism
ko00511 - Other glycan degradation
ko00521 - Streptomycin biosynthesis
ko00540 - Lipopolysaccharide biosynthesis
ko00550 - Peptidoglycan biosynthesis
ko00620 - Pyruvate metabolism
ko00630 - Glyoxylate and dicarboxylate metabolism
ko00633 - Nitrotoluene degradation
ko00640 - Propanoate metabolism
ko00650 - Butanoate metabolism
ko00660 - C5-Branched dibasic acid metabolism
ko00670 - One carbon pool by folate
ko00710 - Carbon fixation in photosynthetic organisms
ko00720 - Carbon fixation pathways in prokaryotes
ko00730 - Thiamine metabolism
ko00740 - Riboflavin metabolism
ko00750 - Vitamin B6 metabolism
ko00760 - Nicotinate and nicotinamide metabolism
ko00770 - Pantothenate and CoA biosynthesis
ko00780 - Biotin metabolism
ko00785 - Lipoic acid metabolism
ko00790 - Folate biosynthesis
ko00900 - Terpenoid backbone biosynthesis
ko00903 - Limonene and pinene degradation
ko00910 - Nitrogen metabolism
ko00970 - Aminoacyl-tRNA biosynthesis
ko00983 - Drug metabolism - other enzymes
ko01040 - Biosynthesis of unsaturated fatty acids
ko01110 - Biosynthesis of secondary metabolites
ko02010 - ABC transporters
ko02020 - Two-component system
ko02030 - Bacterial chemotaxis
ko02040 - Flagellar assembly
ko03010 - Ribosome
ko03018 - RNA degradation
ko03030 - DNA replication
ko03060 - Protein export
ko03070 - Bacterial secretion system
ko03410 - Base excision repair
ko03430 - Mismatch repair
ko03440 - Homologous recombination
ko05111 - Biofilm formation - Vibrio cholerae
M00002 - Glycolysis, core module involving three-carbon compounds
M00003 - Gluconeogenesis, oxaloacetate => fructose-6P
M00005 - PRPP biosynthesis, ribose 5P => PRPP
M00007 - Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P
M00010 - Citrate cycle, first carbon oxidation, oxaloacetate => 2-oxoglutarate
M00012 - Glyoxylate cycle
M00015 - Proline biosynthesis, glutamate => proline
M00016 - Lysine biosynthesis, succinyl-DAP pathway, aspartate => lysine
M00017 - Methionine biosynthesis, apartate => homoserine => methionine
M00018 - Threonine biosynthesis, aspartate => homoserine => threonine
M00019 - Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine
M00020 - Serine biosynthesis, glycerate-3P => serine
M00021 - Cysteine biosynthesis, serine => cysteine
M00022 - Shikimate pathway, phosphoenolpyruvate + erythrose-4P => chorismate
M00027 - GABA (gamma-Aminobutyrate) shunt
M00048 - Inosine monophosphate biosynthesis, PRPP + glutamine => IMP
M00049 - Adenine ribonucleotide biosynthesis, IMP => ADP,ATP
M00050 - Guanine ribonucleotide biosynthesis IMP => GDP,GTP
M00051 - Uridine monophosphate biosynthesis, glutamine (+ PRPP) => UMP
M00053 - Pyrimidine deoxyribonuleotide biosynthesis, CDP/CTP => dCDP/dCTP,dTDP/dTTP
M00060 - Lipopolysaccharide biosynthesis, KDO2-lipid A
M00063 - CMP-KDO biosynthesis
M00064 - ADP-L-glycero-D-manno-heptose biosynthesis
M00086 - beta-Oxidation, acyl-CoA synthesis
M00093 - Phosphatidylethanolamine (PE) biosynthesis, PA => PS => PE
M00096 - C5 isoprenoid biosynthesis, non-mevalonate pathway
M00115 - NAD biosynthesis, aspartate => NAD
M00116 - Menaquinone biosynthesis, chorismate => menaquinol
M00117 - Ubiquinone biosynthesis, prokaryotes, chorismate => ubiquinone
M00119 - Pantothenate biosynthesis, valine/L-aspartate => pantothenate
M00121 - Heme biosynthesis, glutamate => heme
M00123 - Biotin biosynthesis, pimeloyl-ACP/CoA => biotin
M00124 - Pyridoxal biosynthesis, erythrose-4P => pyridoxal-5P
M00126 - Tetrahydrofolate biosynthesis, GTP => THF
M00134 - Polyamine biosynthesis, arginine => ornithine => putrescine
M00140 - C1-unit interconversion, prokaryotes
M00144 - NADH
M00149 - Succinate dehydrogenase, prokaryotes
M00150 - Fumarate reductase, prokaryotes
M00153 - Cytochrome bd ubiquinol oxidase
M00156 - Cytochrome c oxidase, cbb3-type
M00157 - F-type ATPase, prokaryotes and chloroplasts
M00176 - Assimilatory sulfate reduction, sulfate => H2S
M00307 - Pyruvate oxidation, pyruvate => acetyl-CoA
M00364 - C10-C20 isoprenoid biosynthesis, bacteria
M00365 - C10-C20 isoprenoid biosynthesis, archaea
M00432 - Leucine biosynthesis, 2-oxoisovalerate => 2-oxoisocaproate
M00525 - Lysine biosynthesis, acetyl-DAP pathway, aspartate => lysine
M00526 - Lysine biosynthesis, DAP dehydrogenase pathway, aspartate => lysine
M00527 - Lysine biosynthesis, DAP aminotransferase pathway, aspartate => lysine
M00530 - Dissimilatory nitrate reduction, nitrate => ammonia
M00535 - Isoleucine biosynthesis, pyruvate => 2-oxobutanoate
M00549 - Nucleotide sugar biosynthesis, glucose => UDP-glucose
M00565 - Trehalose biosynthesis, D-glucose 1P => trehalose
M00570 - Isoleucine biosynthesis, threonine => 2-oxobutanoate => isoleucine
M00572 - Pimeloyl-ACP biosynthesis, BioC-BioH pathway, malonyl-ACP => pimeloyl-ACP
M00573 - Biotin biosynthesis, BioI pathway, long-chain-acyl-ACP => pimeloyl-ACP => biotin
M00577 - Biotin biosynthesis, BioW pathway, pimelate => pimeloyl-CoA => biotin
M00579 - Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate
M00632 - Galactose degradation, Leloir pathway, galactose => alpha-D-glucose-1P
M00642 - Multidrug resistance, efflux pump MexJK-OprM
M00649 - Multidrug resistance, efflux pump AdeABC
M00705 - Multidrug resistance, efflux pump MepA
M00718 - Multidrug resistance, efflux pump MexAB-OprM
M00793 - dTDP-L-rhamnose biosynthesis
M00840 - Tetrahydrofolate biosynthesis, mediated by ribA and trpF, GTP => THF
M00844 - Arginine biosynthesis, ornithine => arginine
M00845 - Arginine biosynthesis, glutamate => acetylcitrulline => arginine
M00846 - Siroheme biosynthesis, glutamate => siroheme
Undetected
Cell-to-cell spread
Cellular metabolism
Intracellular survival and replication
Invasion
Regulation of gene expression
Stress
Virulence
2-ketobutyrate formate-lyase (EC 2.3.1.-) @ Pyruvate formate-lyase (EC 2.3.1.54) (ID:Z4466)
Carbamoyl-phosphate synthase large chain (EC 6.3.5.5) (ID:Z0038)
D-sedoheptulose 7-phosphate isomerase (EC 5.3.1.28) (ID:SL1344_0306)
GMP synthase [glutamine-hydrolyzing], amidotransferase subunit (EC 6.3.5.2) / GMP synthase [glutamine-hydrolyzing], ATP pyrophosphatase subunit (EC 6.3.5.2) (ID:S2725)
GTP-binding and nucleic acid-binding protein YchF (ID:c1661)
Holliday junction ATP-dependent DNA helicase RuvB (EC 3.6.4.12) (ID:SL1344_1828)
Inosine-5'-monophosphate dehydrogenase (EC 1.1.1.205) / CBS domain (ID:STM2511)
Monothiol glutaredoxin GrxD (ID:E2348C_1740)
Na+/H+-dicarboxylate symporter (ID:Z4942)
NADH-ubiquinone oxidoreductase chain G (EC 1.6.5.3) (ID:STM2323.S)
Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (ID:STM14_5016)
RNA polymerase-binding transcription factor DksA (ID:S0140)
Non-ribosomal peptide synthetase (NRPS)
Other secondary metabolites
MATLAB species model file: msp_0368c.mat