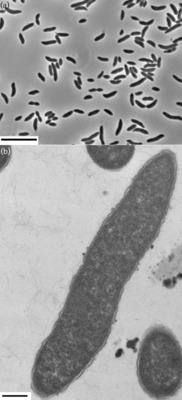
| Species: | unclassified Cloacibacillus |
|---|---|
| Genus: | Cloacibacillus |
| Family: | Synergistaceae |
| Order: | Synergistales |
| Class: | Synergistia |
| Phylum: | Synergistetes |
In the linked pathways:
red=enriched, blue=depleted
ko00010 - Glycolysis / Gluconeogenesis
ko00250 - Alanine, aspartate and glutamate metabolism
ko00290 - Valine, leucine and isoleucine biosynthesis
ko00471 - D-Glutamine and D-glutamate metabolism
ko00473 - D-Alanine metabolism
ko00540 - Lipopolysaccharide biosynthesis
ko00550 - Peptidoglycan biosynthesis
ko00633 - Nitrotoluene degradation
ko00770 - Pantothenate and CoA biosynthesis
ko00780 - Biotin metabolism
ko00970 - Aminoacyl-tRNA biosynthesis
ko03010 - Ribosome
ko03060 - Protein export
ko03070 - Bacterial secretion system
ko03430 - Mismatch repair
M00005 - PRPP biosynthesis, ribose 5P => PRPP
M00019 - Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine
M00031 - Lysine biosynthesis, mediated by LysW, 2-aminoadipate => lysine
M00045 - Histidine degradation, histidine => N-formiminoglutamate => glutamate
M00050 - Guanine ribonucleotide biosynthesis IMP => GDP,GTP
M00060 - Lipopolysaccharide biosynthesis, KDO2-lipid A
M00061 - D-Glucuronate degradation
M00063 - CMP-KDO biosynthesis
M00086 - beta-Oxidation, acyl-CoA synthesis
M00093 - Phosphatidylethanolamine (PE) biosynthesis, PA => PS => PE
M00096 - C5 isoprenoid biosynthesis, non-mevalonate pathway
M00149 - Succinate dehydrogenase, prokaryotes
M00159 - V-type ATPase, prokaryotes
M00308 - Semi-phosphorylative Entner-Doudoroff pathway, gluconate => glycerate-3P
M00432 - Leucine biosynthesis, 2-oxoisovalerate => 2-oxoisocaproate
M00535 - Isoleucine biosynthesis, pyruvate => 2-oxobutanoate
M00565 - Trehalose biosynthesis, D-glucose 1P => trehalose
M00570 - Isoleucine biosynthesis, threonine => 2-oxobutanoate => isoleucine
M00705 - Multidrug resistance, efflux pump MepA
M00844 - Arginine biosynthesis, ornithine => arginine
Undetected
Undetected
Undetected
Undetected
MATLAB species model file: msp_0846.mat