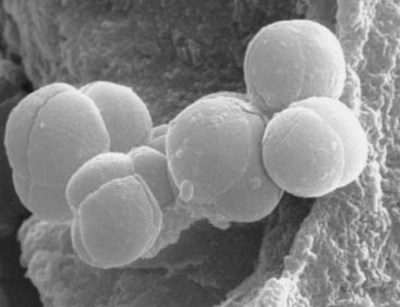
| Species: | Methanosphaera stadtmanae |
|---|---|
| Genus: | Methanosphaera |
| Family: | Methanobacteriaceae |
| Order: | Methanobacteriales |
| Class: | Methanobacteria |
| Phylum: | Euryarchaeota |
| Region Enrichment: | Non-westernized (Fiji, Madagascar, Peru, United Republic of Tanzania) |
|---|
| Shape | Sphere-shaped; Coccoid |
|---|---|
| Gram staining | Gram- |
| Motility | Nonmotile |
| Oxygen Requirement | Anaerobe |
| Sporulation | Nonsporulating |
| Ecosystem | Human; Unclassified |
| Ecosystem Type | Unclassified |
In the linked pathways:
red=enriched, blue=depleted
ko00020 - Citrate cycle (TCA cycle)
ko00072 - Synthesis and degradation of ketone bodies
ko00250 - Alanine, aspartate and glutamate metabolism
ko00290 - Valine, leucine and isoleucine biosynthesis
ko00300 - Lysine biosynthesis
ko00400 - Phenylalanine, tyrosine and tryptophan biosynthesis
ko00471 - D-Glutamine and D-glutamate metabolism
ko00521 - Streptomycin biosynthesis
ko00660 - C5-Branched dibasic acid metabolism
ko00680 - Methane metabolism
ko00730 - Thiamine metabolism
ko00770 - Pantothenate and CoA biosynthesis
ko00860 - Porphyrin and chlorophyll metabolism
ko00970 - Aminoacyl-tRNA biosynthesis
ko03030 - DNA replication
M00005 - PRPP biosynthesis, ribose 5P => PRPP
M00016 - Lysine biosynthesis, succinyl-DAP pathway, aspartate => lysine
M00019 - Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine
M00020 - Serine biosynthesis, glycerate-3P => serine
M00050 - Guanine ribonucleotide biosynthesis IMP => GDP,GTP
M00086 - beta-Oxidation, acyl-CoA synthesis
M00095 - C5 isoprenoid biosynthesis, mevalonate pathway
M00127 - Thiamine biosynthesis, AIR => thiamine-P/thiamine-2P
M00159 - V-type ATPase, prokaryotes
M00356 - Methanogenesis, methanol => methane
M00358 - Coenzyme M biosynthesis
M00364 - C10-C20 isoprenoid biosynthesis, bacteria
M00365 - C10-C20 isoprenoid biosynthesis, archaea
M00378 - F420 biosynthesis
M00432 - Leucine biosynthesis, 2-oxoisovalerate => 2-oxoisocaproate
M00525 - Lysine biosynthesis, acetyl-DAP pathway, aspartate => lysine
M00526 - Lysine biosynthesis, DAP dehydrogenase pathway, aspartate => lysine
M00527 - Lysine biosynthesis, DAP aminotransferase pathway, aspartate => lysine
M00535 - Isoleucine biosynthesis, pyruvate => 2-oxobutanoate
M00567 - Methanogenesis, CO2 => methane
M00570 - Isoleucine biosynthesis, threonine => 2-oxobutanoate => isoleucine
M00608 - 2-Oxocarboxylic acid chain extension, 2-oxoglutarate => 2-oxoadipate => 2-oxopimelate => 2-oxosuberate
M00620 - Incomplete reductive citrate cycle, acetyl-CoA => oxoglutarate
M00705 - Multidrug resistance, efflux pump MepA
M00793 - dTDP-L-rhamnose biosynthesis
M00836 - Coenzyme F430 biosynthesis, sirohydrochlorin => coenzyme F430
M00844 - Arginine biosynthesis, ornithine => arginine
M00845 - Arginine biosynthesis, glutamate => acetylcitrulline => arginine
M00849 - C5 isoprenoid biosynthesis, mevalonate pathway, archaea
Undetected
Undetected
Undetected
Undetected
MATLAB species model file: msp_1311.mat