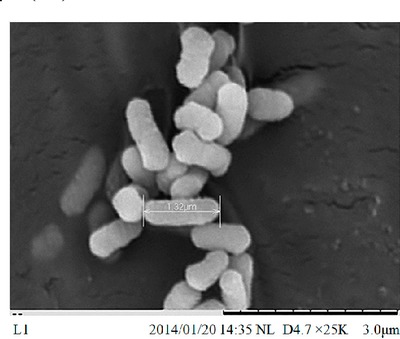
| Species: | Enterobacter cloacae |
|---|---|
| Genus: | Enterobacter |
| Family: | Enterobacteriaceae |
| Order: | Enterobacterales |
| Class: | Gammaproteobacteria |
| Phylum: | Proteobacteria |
| Region Enrichment: | Non-westernized (Fiji, India, Thailand) |
|---|
| Shape | Rod-shaped |
|---|---|
| Gram staining | Gram- |
| Motility | Motile |
| Oxygen Requirement | Facultative; Anaerobe; Aerobe |
| Sporulation | Nonsporulating |
| Ecosystem | Unclassified; Human; Plants; Terrestrial; Mammals; Wastewater; Aquatic |
| Ecosystem Type | Unclassified; Nervous system; Soil; Industrial wastewater; Rhizoplane; Oil reservoir; Circulatory system; Respiratory system; Excretory system; Rhizosphere; Roots |
In the linked pathways:
red=enriched, blue=depleted
ko00010 - Glycolysis / Gluconeogenesis
ko00030 - Pentose phosphate pathway
ko00040 - Pentose and glucuronate interconversions
ko00053 - Ascorbate and aldarate metabolism
ko00061 - Fatty acid biosynthesis
ko00072 - Synthesis and degradation of ketone bodies
ko00130 - Ubiquinone and other terpenoid-quinone biosynthesis
ko00250 - Alanine, aspartate and glutamate metabolism
ko00260 - Glycine, serine and threonine metabolism
ko00270 - Cysteine and methionine metabolism
ko00281 - Geraniol degradation
ko00290 - Valine, leucine and isoleucine biosynthesis
ko00300 - Lysine biosynthesis
ko00410 - beta-Alanine metabolism
ko00450 - Selenocompound metabolism
ko00471 - D-Glutamine and D-glutamate metabolism
ko00473 - D-Alanine metabolism
ko00480 - Glutathione metabolism
ko00500 - Starch and sucrose metabolism
ko00520 - Amino sugar and nucleotide sugar metabolism
ko00521 - Streptomycin biosynthesis
ko00540 - Lipopolysaccharide biosynthesis
ko00550 - Peptidoglycan biosynthesis
ko00620 - Pyruvate metabolism
ko00670 - One carbon pool by folate
ko00710 - Carbon fixation in photosynthetic organisms
ko00730 - Thiamine metabolism
ko00750 - Vitamin B6 metabolism
ko00770 - Pantothenate and CoA biosynthesis
ko00780 - Biotin metabolism
ko00785 - Lipoic acid metabolism
ko00791 - Atrazine degradation
ko00903 - Limonene and pinene degradation
ko00910 - Nitrogen metabolism
ko00983 - Drug metabolism - other enzymes
ko02010 - ABC transporters
ko02030 - Bacterial chemotaxis
ko02040 - Flagellar assembly
ko02060 - Phosphotransferase system (PTS)
ko03430 - Mismatch repair
ko03440 - Homologous recombination
M00007 - Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P
M00008 - Entner-Doudoroff pathway, glucose-6P => glyceraldehyde-3P + pyruvate
M00015 - Proline biosynthesis, glutamate => proline
M00016 - Lysine biosynthesis, succinyl-DAP pathway, aspartate => lysine
M00017 - Methionine biosynthesis, apartate => homoserine => methionine
M00060 - Lipopolysaccharide biosynthesis, KDO2-lipid A
M00061 - D-Glucuronate degradation
M00063 - CMP-KDO biosynthesis
M00086 - beta-Oxidation, acyl-CoA synthesis
M00093 - Phosphatidylethanolamine (PE) biosynthesis, PA => PS => PE
M00096 - C5 isoprenoid biosynthesis, non-mevalonate pathway
M00116 - Menaquinone biosynthesis, chorismate => menaquinol
M00117 - Ubiquinone biosynthesis, prokaryotes, chorismate => ubiquinone
M00123 - Biotin biosynthesis, pimeloyl-ACP/CoA => biotin
M00124 - Pyridoxal biosynthesis, erythrose-4P => pyridoxal-5P
M00127 - Thiamine biosynthesis, AIR => thiamine-P/thiamine-2P
M00134 - Polyamine biosynthesis, arginine => ornithine => putrescine
M00150 - Fumarate reductase, prokaryotes
M00153 - Cytochrome bd ubiquinol oxidase
M00338 - Cysteine biosynthesis, homocysteine + serine => cysteine
M00364 - C10-C20 isoprenoid biosynthesis, bacteria
M00365 - C10-C20 isoprenoid biosynthesis, archaea
M00525 - Lysine biosynthesis, acetyl-DAP pathway, aspartate => lysine
M00526 - Lysine biosynthesis, DAP dehydrogenase pathway, aspartate => lysine
M00527 - Lysine biosynthesis, DAP aminotransferase pathway, aspartate => lysine
M00530 - Dissimilatory nitrate reduction, nitrate => ammonia
M00550 - Ascorbate degradation, ascorbate => D-xylulose-5P
M00554 - Nucleotide sugar biosynthesis, galactose => UDP-galactose
M00555 - Betaine biosynthesis, choline => betaine
M00565 - Trehalose biosynthesis, D-glucose 1P => trehalose
M00572 - Pimeloyl-ACP biosynthesis, BioC-BioH pathway, malonyl-ACP => pimeloyl-ACP
M00573 - Biotin biosynthesis, BioI pathway, long-chain-acyl-ACP => pimeloyl-ACP => biotin
M00577 - Biotin biosynthesis, BioW pathway, pimelate => pimeloyl-CoA => biotin
M00631 - D-Galacturonate degradation (bacteria)
M00632 - Galactose degradation, Leloir pathway, galactose => alpha-D-glucose-1P
M00641 - Multidrug resistance, efflux pump MexEF-OprN
M00642 - Multidrug resistance, efflux pump MexJK-OprM
M00649 - Multidrug resistance, efflux pump AdeABC
M00696 - Multidrug resistance, efflux pump AcrEF-TolC
M00704 - Tetracycline resistance, efflux pump Tet38
M00718 - Multidrug resistance, efflux pump MexAB-OprM
M00745 - Imipenem resistance, repression of porin OprD
M00761 - Undecaprenylphosphate alpha-L-Ara4N biosynthesis, UDP-GlcA => undecaprenyl phosphate alpha-L-Ara4N
M00793 - dTDP-L-rhamnose biosynthesis
M00843 - L-threo-Tetrahydrobiopterin biosynthesis, GTP => L-threo-BH4
Biofilm
Cell motility
Cell surface and membrane proteins
Cell-to-cell spread
Cellular metabolism
Chaperone
Drug efflux system
Fimbriae
Intracellular survival and replication
Invasion
Lps (lipopolysaccharide)
Regulation of gene expression
Two-component system
Virulence
Zinc uptake
16 kDa heat shock protein B (ID:c4606)
3'-to-5' exoribonuclease RNase R (ID:S4602)
4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase (EC 5.3.1.17) (ID:STM474_3165)
ATP-dependent helicase HrpA (ID:Z2313)
Biosynthetic arginine decarboxylase (EC 4.1.1.19) (ID:Z4283)
Carbon starvation protein A (ID:SL1344_0588)
Chorismate synthase (EC 4.2.3.5) (ID:t0480)
Colanic acid biosynthesis glycosyl transferase WcaE (ID:STM474_2196)
Curli production assembly/transport component CsgG (ID:STM14_1303)
D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (ID:Z4251)
D-tagatose-1,6-bisphosphate aldolase subunit KbaY (EC 4.1.2.40) (ID:b3137)
Deoxyguanosinetriphosphate triphosphohydrolase (EC 3.1.5.1), subgroup 1 (ID:SL1344_0209)
DNA polymerase IV (EC 2.7.7.7) (ID:SL1344_0309)
DTDP-4-amino-4,6-dideoxygalactose transaminase (EC 2.6.1.59) (ID:c4711)
Galactose/methyl galactoside ABC transporter, ATP-binding protein MglA (EC 3.6.3.17) (ID:Z3404)
GMP synthase [glutamine-hydrolyzing], amidotransferase subunit (EC 6.3.5.2) / GMP synthase [glutamine-hydrolyzing], ATP pyrophosphatase subunit (EC 6.3.5.2) (ID:S2725)
Hha toxicity modulator TomB (ID:STM474_0494)
Holliday junction ATP-dependent DNA helicase RuvB (EC 3.6.4.12) (ID:SL1344_1828)
Homoserine O-succinyltransferase (EC 2.3.1.46) (ID:SL1344_4117)
Inactive (p)ppGpp 3'-pyrophosphohydrolase domain / GTP pyrophosphokinase (EC 2.7.6.5), (p)ppGpp synthetase I (ID:SG2866)
Lipid A biosynthesis myristoyltransferase (EC 2.3.1.243) (ID:SFK218_2097)
Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) (ID:SL1344_1747)
Manganese ABC transporter, ATP-binding protein SitB (ID:c1599)
Multidrug efflux pump MdfA/Cmr (of MFS type), broad spectrum (ID:STM14_1016)
Murein peptide ABC transporter, substrate-binding protein (requires DppBCDF) (ID:c1803)
Na+/H+-dicarboxylate symporter (ID:Z4942)
Osmolarity sensory histidine kinase EnvZ (ID:STM14_4216)
Outer membrane porin OmpD (ID:SL1344_1503)
Outer-membrane-phospholipid-binding lipoprotein MlaA (ID:S2559)
Oxygen-insensitive NAD(P)H nitroreductase (EC 1.-.-.-) / Dihydropteridine reductase (EC 1.5.1.34) (ID:SL1344_0566)
Periplasmic beta-glucosidase (EC 3.2.1.21) (ID:SL1344_2144)
Periplasmic chaperone and peptidyl-prolyl cis-trans isomerase of outer membrane proteins SurA (EC 5.2.1.8) (ID:SF0050)
Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (ID:STM14_5016)
Probable endopeptidase NlpC (ID:SL1344_1277)
SbmA protein (ID:c0482)
Sensor histidine kinase PhoQ (EC 2.7.13.3) (ID:STM14_1408)
Signal transduction histidine-protein kinase BarA (EC 2.7.13.3) (ID:SL1344_2939)
Thioredoxin 2 (ID:Z3867)
Threonine catabolic operon transcriptional activator TdcA (ID:SL1344_3217)
TonB-dependent receptor; Outer membrane receptor for ferric enterobactin and colicins B, D (ID:STM14_0682)
Trk potassium uptake system protein TrkA (ID:Z4660)
UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase (EC 2.3.1.191) (ID:STM0226)
UDP-glucose 6-dehydrogenase (EC 1.1.1.22) (ID:SL1344_2057)
Uncharacterized HTH-type transcriptional regulator YfhH (ID:Z3841)
Uncharacterized protease YegQ (ID:SL1344_2112)
Undecaprenyl-phosphate alpha-N-acetylglucosaminyl 1-phosphate transferase (EC 2.7.8.33) (ID:S3902)
UPF0187 protein YneE (ID:STM474_1538)
Zinc ABC transporter, ATP-binding protein ZnuC (ID:STM14_2301)
Zinc ABC transporter, permease protein ZnuB (ID:STM14_2302)
Zinc ABC transporter, substrate-binding protein ZnuA (ID:STM14_2300)
Aryl Polyene
Non-ribosomal peptide synthetase (NRPS)
Siderophore
Type-II PKS-Ladderane